Give an account of Hershey and Chase experiment. What did it conclusively prove? If both DNA and proteins contained phosphorus and sulphur do you think the result would have been the same?
Hershey and Chase conducted experiments on bacteriophage to prove that DNA is the genetic material.


 Hershey and Chase experiment
Hershey and Chase experiment
(i) Some bacteriophage virus were grown on a medium that contained radioactive phosphorus ( ${ }^{32} \mathrm{P}$ ) and some in another medium with radioactive sulphur ( ${ }^{35} \mathrm{~S}$ ).
(ii) Viruses grown in the presence of radioactive phosphorus ( ${ }^{32} \mathrm{P}$ ) contained radioactive DNA.
(iii) Similar viruses grown in presence of radioactive sulphur ( ${ }^{35} \mathrm{~S}$ ) contained radioactive protein.
(iv) Both the radioactive virus types were allowed to infect E. coli separately.
(v) Soon after infection, the bacterial cells were gently agitated in blender to remove viral coats from the bacteria.
(vi) The culture was also centrifuged to separate the viral particle from the bacterial cell.
Observations and Conclusions
(i) Only radioactive ${ }^{32} \mathrm{P}$ was found to be associated with the bacterial cell, whereas radioactive ${ }^{35} \mathrm{~S}$ was only found in surrounding medium and not in the bacterial cell.
(ii) This indicates that only DNA and not protein coat entered the bacterial cell.
(iii) This proves that DNA is the genetic material which is passed from virus to bacteria and not protein.
If both DNA and proteins contained phosphorus and sulphur, the result might change.
In case (i)
Radioactive ${ }^{35} \mathrm{~S}$ and + Bacteriophage ${ }^{32} \mathrm{P}$ labelled protein capsule $\longrightarrow$ No radioactive
${ }^{35} \mathrm{~S}$ and ${ }^{32} \mathrm{P}$ Detected in cells + Radioactivity ( ${ }^{35} \mathrm{~S}$ and ${ }^{32} \mathrm{P}$ ) detected in supernatant
In case (ii)
Radioactive ${ }^{35} \mathrm{~S}$ and ${ }^{32} \mathrm{P}$ lebelled DNA + Bacteriophage $\longrightarrow$ Radioactive ${ }^{32} \mathrm{P}$ and ${ }^{35} \mathrm{~S}$
Detected in cells + No radioactivity detected in supernatant
During the course of evolution why DNA was choosen over RNA as genetic material. Give reasons by first discussing the desired criteria in a molecule that can act as genetic material and in the light of biochemical differences between DNA and RNA.
A molecule that can act as a genetic material must fulfil the following
(i) It should be able to generate its replica (replication).
(ii) It should chemically and structurally be stable.
(iii) It should provide the scope for slow changes (mutation) that are required for evolution.
(iv) It should be able to express itself in the form of Mendelian.
Biochemical differences between DNA and RNA
(i) Both nucleic acid (DNA and RNA) are able to direct their duplication proteins fails for the first criteria.
(ii) RNA is reactive, it also acts are catalyst, hence DNA is less reactive and structurally more stable than RNA.
(iii) Presence of thymine at the place of uracil also confers additional stability to DNA.
Give an account of post transcriptional modifications of a eukaryotic mRNA.
Post-transcriptional Modifications
The primary transcripts are non-functional, containing both the coding region, exon and non-coding region, intron in RNA and are called heterogenous RNA or hnRNA. In eukaryotes, three types of RNA polymerases are found in the nucleus
(i) RNA polymerase I transcribes rRNAs ( 28 S and 5.8 S ).
(ii) RNA polymerase II transcribes the precursor of mRNA (called heterogeneous nuclear RNA or hnRNA).
(iii) RNA polymerase III transcribes $t$ RNA, 5 S rRNA and snRNAs (small nuclear RNAs).
The hnRNA undergoes two additional processes called capping and tailing.
In capping, an unusual nucleotide, methyl guanosine triphosphate is added to the 5 '-end of hnRNA.
In tailing, adenylate residues (about 200-300) are added at 3'-end in a template independent manner.
Now the hnRNA undergoes a process where the introns are removed and exons are joined to form $m$ RNA by the process called splicing.
 $$ \text { Diagram representation of a post transcriptional modification in eukaryotes } $$
$$ \text { Diagram representation of a post transcriptional modification in eukaryotes } $$
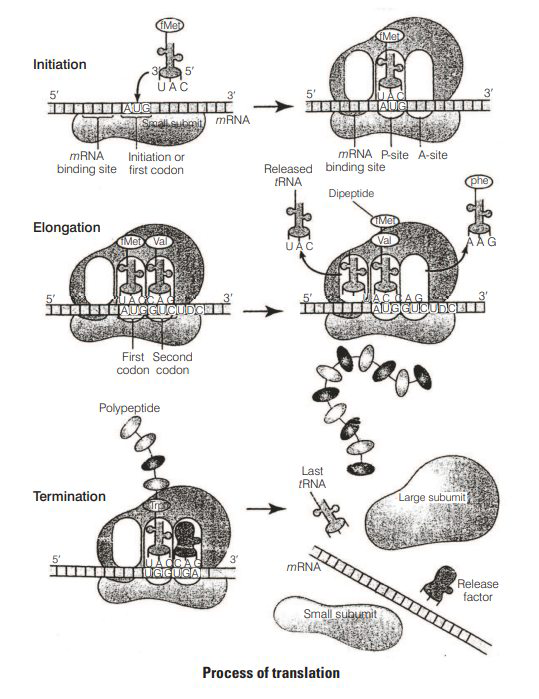
$$ \text { Discuss the process of translation in detail. } $$
There are three-stages of protein synthesis
(i) Initiation
Assembly of Ribosomes on mRNA In prokaryotes, initiation requires the large and small ribosome subunits, the mRNA, initiation $t$ RNA and three Initiation Factors (IFs).
Activation of Amino Acid Amino acids become activated by binding with aminoacyl $t$ RNA synthetase enzyme in the presence of ATP.
$$ \text { Amino acid (AA) }+ \text { ATP } \xrightarrow[\text { synthetases }]{\text { AminoacytRNA }} \text { AA-AMP-Enzyme complex }+P_i $$
Transfer of Amino Acid to tRNA The AA-AMP-enzyme complex formed reacts with specific $t$ RNA to form aminoacyl $t$ RNA complex.
$$ \text { AA-AMP-Enzyme complex }+t \text { RNA } \longrightarrow \text { AA } t \text { RNA }+ \text { AMP }+ \text { Enzyme. } $$
The cap region of $m R N A$ binds to the smaller subunit of ribosome.
The ribosome has two sites, A-site and P-site.
The smaller subunit first binds the initiator $t$ RNA then and then binds to the larger subunit so, that initiation codon (AUG) lies on the P-site.
The initiation $t$ RNA, i.e., methionyl $t$ RNA then binds to the P -site.
(ii) Elongation
Another charged aminoacyltRNA complex binds to the A-site of the ribosome.
Peptide bond formation and movement along the mRNA called translocation. A peptide bond is formed between carboxyl group ( -COOH ) of amino acid at P -site and amino group ( -NH ) of amino acid at A-site by the enzyme peptidyl transferase.
The ribosome slides over $m R N A$ from codon to codon in the $5^{\prime} \rightarrow 3^{\prime}$ direction.
According to the sequence of codon, amino acids are attached to one another by peptide bonds and a polypeptide chain is formed.
(iii) Termination
When the A-site of ribosome reaches a termination codon which does not code for any amino acid, no charged $t$ RNA binds to the A-site.
Dissociation of polypeptide from ribosome takes place which is catalysed by a 'release factor'.
There are three termination codons, i.e., UGA, UAG and UAA.
Define an operon, giving an example, explain an inducible operon.
The concept of operon was first proposed in 1961, by Jacob and Monad. An operon is a unit of prokaryotic gene expression which includes coordinately regulated (structural) genes and control elements which are recognised by regulatory gene product.
Components of an Operon
(i) Structural gene The fragment of DNA which transcribe mRNA for polypeptide synthesis.
(ii) Promoter The sequence of DNA where RNA polymerase binds and initiates transcription of structural genes is called promoter.
(iii) Operator The sequence of DNA adjacent to promoter where specific repressor protein binds is called operator.
(iv) Regulator gene The gene that codes for the repressor protein that binds to the operator and suppresses its activity as a result of which transcription will be switched off.
(v) Inducer The substrate that prevents the repressor from binding to the operator, is called an inducer. As a result transcription is switched on. It is a chemical of diverse nature like metabolite, hormone substrate, etc.
Inducible Operon System
An inducible operon system is a regulated unit of genetic material which is switched on in response to the presence of a chemical. e.g., the lactose or lac-operon of E.coli.
The lactose operon The lac $z, y$, a genes are transcribed from a lac transcription unit under the control of a single promoter. They encode enzyme required for the use of lactose as a carbon source. The lac $i$ gene product, the lac repressor, is expressed from a separate transcription unit upstream from the operator.
lac operon consists of three structural genes ( $z, y$ and a), operator, promoter and a separate regulatory gene.
The three structural genes $(a, y$ and $a)$ transcribe a polycistronic $m R N A$.
 Lac operon
Lac operon
Gene $z$ codes for $\beta$-galactosidase ( $\beta$-gal) enzyme which breaks lactose into galactose and glucose.
Gene $y$ codes for permease, which increases the permeability of the cell to lactose.
Gene a codes for enzyme transacetylase, which catalyses the transacetylation of lactose in its active form.
When Lactose is Absent
(i) When lactose is absent, $i$ gene regulates and produces repressor mRNA which translate repression.
(ii) The repressor protein binds to the operator region of the operon and as a result prevents RNA polymerase to bind to the operon.
(iii) The operon is switched off.
When Lactose is Present
(i) Lactose acts as an inducer which binds to the repressor and forms an inactive repressor.
(ii) The repressor fails to bind to the operator region.
(iii) The RNA polymerase binds to the operator and transcript lac mRNA.
(iv) lac mRNA is polycistronic, i.e., produces all three enzymes, $\beta$-galactosidase, permease and transacetylase.
(v) The lac operon is switched on.