$$ \text { Discuss the process of translation in detail. } $$
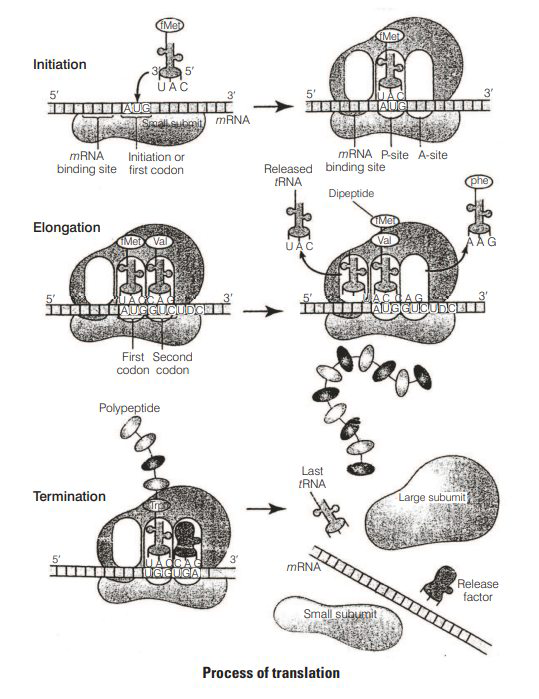
There are three-stages of protein synthesis
(i) Initiation
Assembly of Ribosomes on mRNA In prokaryotes, initiation requires the large and small ribosome subunits, the mRNA, initiation $t$ RNA and three Initiation Factors (IFs).
Activation of Amino Acid Amino acids become activated by binding with aminoacyl $t$ RNA synthetase enzyme in the presence of ATP.
$$ \text { Amino acid (AA) }+ \text { ATP } \xrightarrow[\text { synthetases }]{\text { AminoacytRNA }} \text { AA-AMP-Enzyme complex }+P_i $$
Transfer of Amino Acid to tRNA The AA-AMP-enzyme complex formed reacts with specific $t$ RNA to form aminoacyl $t$ RNA complex.
$$ \text { AA-AMP-Enzyme complex }+t \text { RNA } \longrightarrow \text { AA } t \text { RNA }+ \text { AMP }+ \text { Enzyme. } $$
The cap region of $m R N A$ binds to the smaller subunit of ribosome.
The ribosome has two sites, A-site and P-site.
The smaller subunit first binds the initiator $t$ RNA then and then binds to the larger subunit so, that initiation codon (AUG) lies on the P-site.
The initiation $t$ RNA, i.e., methionyl $t$ RNA then binds to the P -site.
(ii) Elongation
Another charged aminoacyltRNA complex binds to the A-site of the ribosome.
Peptide bond formation and movement along the mRNA called translocation. A peptide bond is formed between carboxyl group ( -COOH ) of amino acid at P -site and amino group ( -NH ) of amino acid at A-site by the enzyme peptidyl transferase.
The ribosome slides over $m R N A$ from codon to codon in the $5^{\prime} \rightarrow 3^{\prime}$ direction.
According to the sequence of codon, amino acids are attached to one another by peptide bonds and a polypeptide chain is formed.
(iii) Termination
When the A-site of ribosome reaches a termination codon which does not code for any amino acid, no charged $t$ RNA binds to the A-site.
Dissociation of polypeptide from ribosome takes place which is catalysed by a 'release factor'.
There are three termination codons, i.e., UGA, UAG and UAA.
Define an operon, giving an example, explain an inducible operon.
The concept of operon was first proposed in 1961, by Jacob and Monad. An operon is a unit of prokaryotic gene expression which includes coordinately regulated (structural) genes and control elements which are recognised by regulatory gene product.
Components of an Operon
(i) Structural gene The fragment of DNA which transcribe mRNA for polypeptide synthesis.
(ii) Promoter The sequence of DNA where RNA polymerase binds and initiates transcription of structural genes is called promoter.
(iii) Operator The sequence of DNA adjacent to promoter where specific repressor protein binds is called operator.
(iv) Regulator gene The gene that codes for the repressor protein that binds to the operator and suppresses its activity as a result of which transcription will be switched off.
(v) Inducer The substrate that prevents the repressor from binding to the operator, is called an inducer. As a result transcription is switched on. It is a chemical of diverse nature like metabolite, hormone substrate, etc.
Inducible Operon System
An inducible operon system is a regulated unit of genetic material which is switched on in response to the presence of a chemical. e.g., the lactose or lac-operon of E.coli.
The lactose operon The lac $z, y$, a genes are transcribed from a lac transcription unit under the control of a single promoter. They encode enzyme required for the use of lactose as a carbon source. The lac $i$ gene product, the lac repressor, is expressed from a separate transcription unit upstream from the operator.
lac operon consists of three structural genes ( $z, y$ and a), operator, promoter and a separate regulatory gene.
The three structural genes $(a, y$ and $a)$ transcribe a polycistronic $m R N A$.
 Lac operon
Lac operon
Gene $z$ codes for $\beta$-galactosidase ( $\beta$-gal) enzyme which breaks lactose into galactose and glucose.
Gene $y$ codes for permease, which increases the permeability of the cell to lactose.
Gene a codes for enzyme transacetylase, which catalyses the transacetylation of lactose in its active form.
When Lactose is Absent
(i) When lactose is absent, $i$ gene regulates and produces repressor mRNA which translate repression.
(ii) The repressor protein binds to the operator region of the operon and as a result prevents RNA polymerase to bind to the operon.
(iii) The operon is switched off.
When Lactose is Present
(i) Lactose acts as an inducer which binds to the repressor and forms an inactive repressor.
(ii) The repressor fails to bind to the operator region.
(iii) The RNA polymerase binds to the operator and transcript lac mRNA.
(iv) lac mRNA is polycistronic, i.e., produces all three enzymes, $\beta$-galactosidase, permease and transacetylase.
(v) The lac operon is switched on.
'There is a paternity dispute for a child'. Which technique can solve the problem? Discuss the principle involved.
DNA fingerprinting is the technique used in solving the paternity dispute for a child. DNA fingerprinting is a technique of determining nucleotide sequences of certain areas of DNA which are unique to each individual.
The basis of DNA fingerprinting is DNA polymorphism. Although the DNA from different individuals is more alike than different, there are many regions of the human chromosomes that exhibit a great deal of diversity. Such variable sequences are termed 'polymorphic' (meaning many forms).
A special type of polymorphism, called VNTR (Variable Number of Tandem Repeats), is composed of repeated copies of a DNA sequence that lie adjacent to one another on the chromosome. Since, polymorphism is the basis of genetic mapping of human.
Give an account of the methods used in sequencing the human genome.
Sequencing of human genome has made it possible to understand the link between various genes and their functions.
If there are any gene defects that express as disorders or that increase the susceptibility of an individual to a disease then specific gene therapies can be worked out
Methodologies of human genome sequencing
The methods involve two major approaches
(i) Expressed Sequence Tags (ESTs) This method focusses on identifying all the genes that are expressed as RNA.
(ii) Sequence annotation It is an approach of simply sequencing the whole set of genome that contains all the coding and non-coding sequences, and later assigning different regions in the sequence with functions.
For sequencing, first the total DNA from cell is i.e., solated and broken down in relatively small sizes as fragments.
There DNA fragments are cloned in suitable host using suitable vectors. When bacteria is used as vector, they are called Bacterial Artificial Chromosomes (BAC) and when yeast is used as vector, they are called Yeast Artificial Chromosomes (YACs).
Frederick Sanger developed a principle according to which the fragments of DNA are sequenced by automated DNA sequences.
On the basis of overlapping regions on DNA fragments, these sequences are arranged accordingly. For alignment of these sequences, specialised computer-based programmes were developed.
Finally, the genetic and physical maps of the genome were constructed by collecting information about certain repetitive DNA sequences and DNA polymorphism, based on endonuclease recognition sites.
Dr. Alec Jeffreys developed the technique of DNA fingerprinting in an attempt to identify DNA marker for inherited diseases.
DNA fingerprinting uses short nucleotide repeats called Variable Number Tandem Repeats (VNTRs) as markers. VNTRs vary from person to person and are inherited from one generation to the next. Only closely individuals have similar VNTRs.